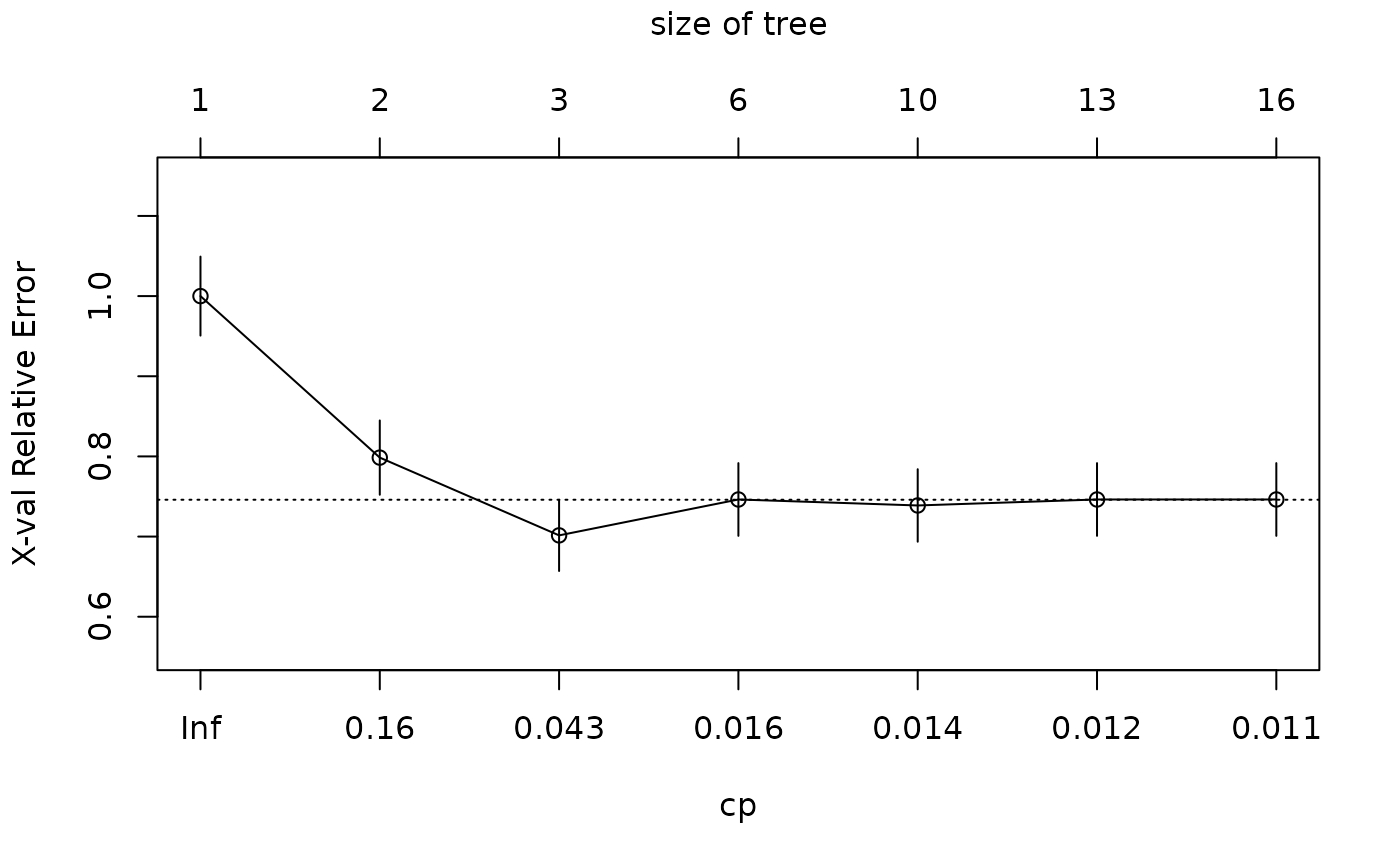
Complexity Parameter of an rpart Model
CP.RdExtracts, prints and plots the complexity table of an rpart model.
Arguments
- x
fitted model object of class
"rpart". This is assumed to be the result of some function that produces an object with the same named components as that returned by therpartfunction.- digits
the number of digits of numbers to print.
- minline
whether a horizontal line is drawn 1SE above the minimum of the curve.
- lty
line type for this line
- col
colour for this line
- upper
what is plotted on the top axis: the size of the tree (the number of leaves) ("
size"), the number of splits ("splits") or nothing ("none").- ...
further arguments passed to
printandplot
Details
The complexity parameter table is hidden deep in the entrails of the rpart result object, it is convenient to have a function to extract it.
Examples
r.rp <- FitMod(diabetes ~ ., d.pima, fitfn="rpart")
CP(r.rp)
#>
#> Classification tree:
#> rpart(formula = diabetes ~ ., data = d.pima, model = TRUE, y = TRUE)
#>
#> Variables actually used in tree construction:
#> [1] age glucose insulin mass pedigree pregnant pressure
#>
#> Root node error: 268/768 = 0.34896
#>
#> n= 768
#>
#> CP nsplit rel error xerror xstd
#> 1 0.242537 0 1.00000 1.00000 0.049288
#> 2 0.104478 1 0.75746 0.79851 0.046360
#> 3 0.017413 2 0.65299 0.70149 0.044461
#> 4 0.014925 5 0.60075 0.74627 0.045381
#> 5 0.013060 9 0.54104 0.73881 0.045233
#> 6 0.011194 12 0.49254 0.74627 0.045381
#> 7 0.010000 15 0.45896 0.74627 0.045381
#>
plot(CP(r.rp))