Finds many (all) roots of one equation within an interval
UnirootAll.RdThe function UnirootAll searches the interval from lower to upper
for several roots (i.e., zero's) of a function f with respect to
its first argument.
Arguments
- f
the function for which the root is sought.
- interval
a vector containing the end-points of the interval to be searched for the root.
- lower
the lower end point of the interval to be searched.
- upper
the upper end point of the interval to be searched.
- tol
the desired accuracy (convergence tolerance).
- maxiter
the maximum number of iterations.
- n
number of subintervals in which the root is sought.
- ...
additional named or unnamed arguments to be passed to
f(but beware of partial matching to other arguments).
Note
This is a verbatim copy from rootSolve::uniroot.all (v. 1.7).
Value
a vector with the roots found in the interval
Details
f will be called as f(x, ...) for a numeric value of x.
Run demo(Jacobandroots) for an example of the use of UnirootAll
for steady-state analysis.
See also second example of gradient
This example is discussed in the book by Soetaert and Herman (2009).
See also
uniroot for more information about input.
Note
The function calls uniroot, the basic R-function.
It is not guaranteed that all roots will be recovered.
This will depend on n, the number of subintervals in which the
interval is divided.
If the function "touches" the X-axis (i.e. the root is a saddle point), then this root will generally not be retrieved. (but chances of this are pretty small).
Whereas unitroot passes values one at a time to the function,
UnirootAll passes a vector of values to the function.
Therefore f should be written such that it can handle a vector of values.
See last example.
Examples
## =======================================================================
## Mathematical examples
## =======================================================================
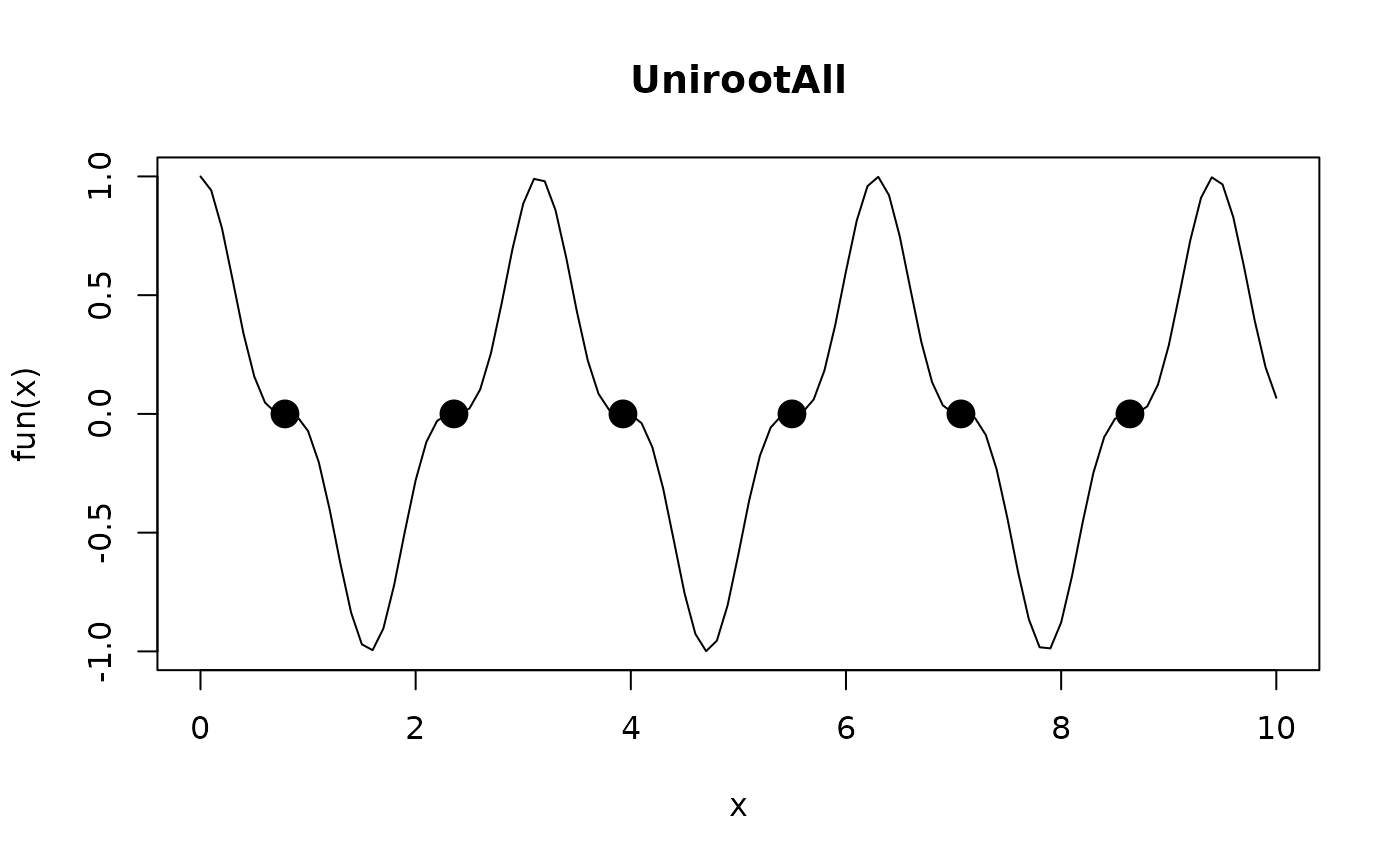
# a well-behaved case...
fun <- function (x) cos(2*x)^3
curve(fun(x), 0, 10,main = "UnirootAll")
All <- UnirootAll(fun, c(0, 10))
points(All, y = rep(0, length(All)), pch = 16, cex = 2)
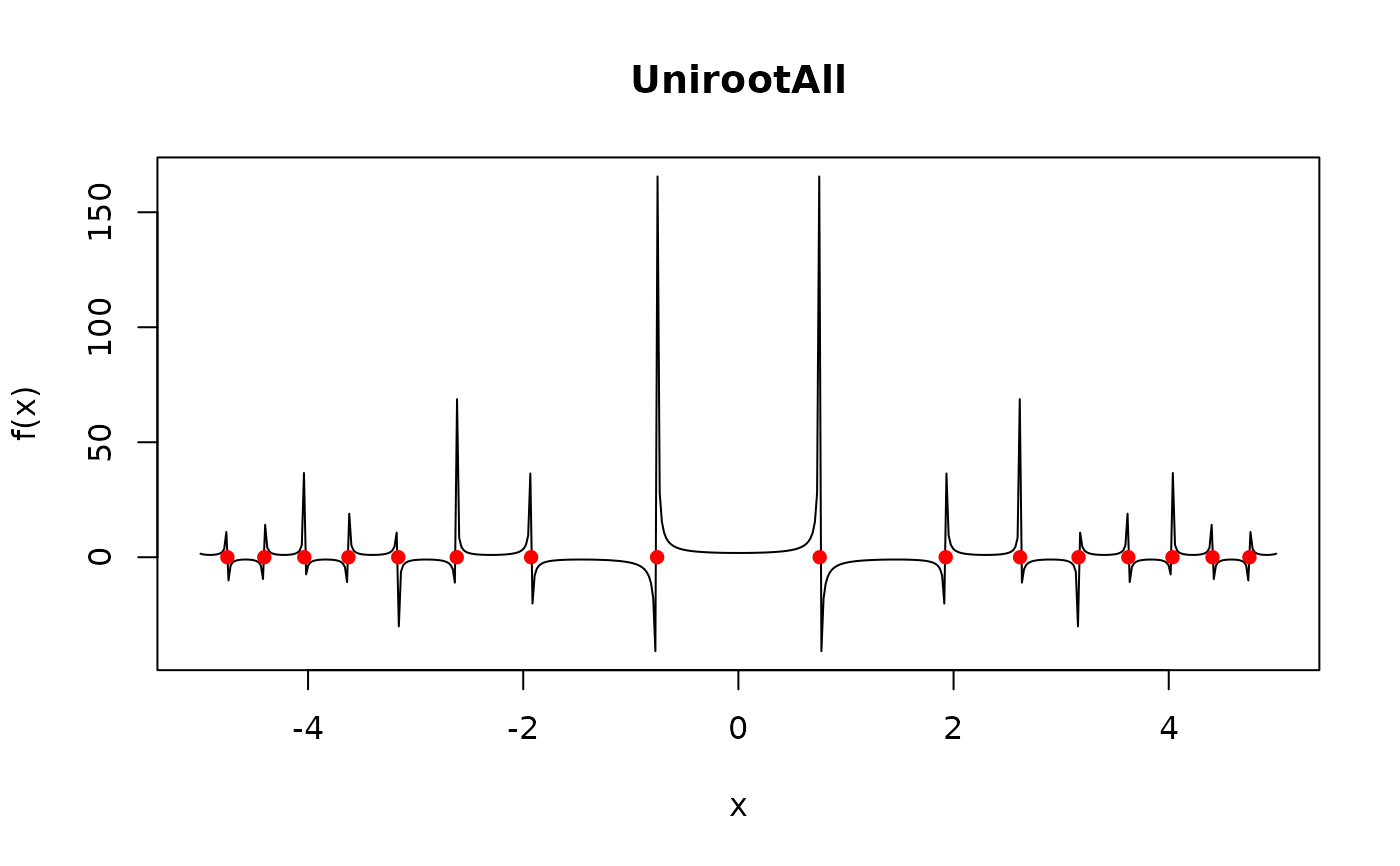
 # a difficult case...
f <- function (x) 1/cos(1+x^2)
AA <- UnirootAll(f, c(-5, 5))
curve(f(x), -5, 5, n = 500, main = "UnirootAll")
points(AA, rep(0, length(AA)), col = "red", pch = 16)
# a difficult case...
f <- function (x) 1/cos(1+x^2)
AA <- UnirootAll(f, c(-5, 5))
curve(f(x), -5, 5, n = 500, main = "UnirootAll")
points(AA, rep(0, length(AA)), col = "red", pch = 16)
 f(AA) # !!!
#> [1] -19783385 10048135 -17778378 17716401 19705646 21300284 -61531208
#> [8] -52150718 -52150685 -61531329 21300313 19705621 17716401 -17778378
#> [15] 10048135 -19783436
## =======================================================================
## Vectorisation:
## =======================================================================
# from R-help Digest, Vol 130, Issue 27
# https://stat.ethz.ch/pipermail/r-help/2013-December/364799.html
integrand1 <- function(x) 1/x*dnorm(x)
integrand2 <- function(x) 1/(2*x-50)*dnorm(x)
integrand3 <- function(x, C) 1/(x+C)
res <- function(C) {
integrate(integrand1, lower = 1, upper = 50)$value +
integrate(integrand2, lower = 50, upper = 100)$value -
integrate(integrand3, C = C, lower = 1, upper = 100)$value
}
# uniroot passes one value at a time to the function, so res can be used as such
uniroot(res, c(1, 1000))
#> $root
#> [1] 837
#>
#> $f.root
#> [1] 0.0000000000162
#>
#> $iter
#> [1] 9
#>
#> $init.it
#> [1] NA
#>
#> $estim.prec
#> [1] 0.000061
#>
# Need to vectorise the function to use UnirootAll:
res <- Vectorize(res)
UnirootAll(res, c(1,1000))
#> [1] 837
f(AA) # !!!
#> [1] -19783385 10048135 -17778378 17716401 19705646 21300284 -61531208
#> [8] -52150718 -52150685 -61531329 21300313 19705621 17716401 -17778378
#> [15] 10048135 -19783436
## =======================================================================
## Vectorisation:
## =======================================================================
# from R-help Digest, Vol 130, Issue 27
# https://stat.ethz.ch/pipermail/r-help/2013-December/364799.html
integrand1 <- function(x) 1/x*dnorm(x)
integrand2 <- function(x) 1/(2*x-50)*dnorm(x)
integrand3 <- function(x, C) 1/(x+C)
res <- function(C) {
integrate(integrand1, lower = 1, upper = 50)$value +
integrate(integrand2, lower = 50, upper = 100)$value -
integrate(integrand3, C = C, lower = 1, upper = 100)$value
}
# uniroot passes one value at a time to the function, so res can be used as such
uniroot(res, c(1, 1000))
#> $root
#> [1] 837
#>
#> $f.root
#> [1] 0.0000000000162
#>
#> $iter
#> [1] 9
#>
#> $init.it
#> [1] NA
#>
#> $estim.prec
#> [1] 0.000061
#>
# Need to vectorise the function to use UnirootAll:
res <- Vectorize(res)
UnirootAll(res, c(1,1000))
#> [1] 837